Using the dataRetrieval Stats Service
Using the R package dataRetrieval Stats Service.
Introduction
This script utilizes the new dataRetrieval package access to the USGS Statistics Web Service
. We will be pulling daily mean data using the daily value service in readNWISdata, and using the stats service data to put it in the context of the site’s history. Here we are retrieving data for July 12th in the Upper Midwest, where a major storm system had recently passed through. You can modify this script to look at other areas and dates simply by modifying the states and storm.date objects.
To run this code, we recommend having either dataRetreival version 2.5.13 (currently the latest release on CRAN) or version 2.6.1 (currently the latest Github release).
Get the data
There are two separate dataRetrieval calls here — one to retrieve the daily discharge data, and one to retrieve the historical discharge statistics. Both calls are inside loops to split them into smaller pieces, to accomodate web service restrictions. The daily values service allows only single states as a filter, so we loop over the list of states. The stats service does not allow requests of more than ten sites, so the loop iterates by groups of ten site codes. Retrieving the data can take a few tens of seconds. Once we have both the daily value and statistics data, the two data frames are joined by site number via dplyr’s
left_join function. We use a pipe
to send the output of the join to na.omit() function. Then we add a column to the final data frame to hold the color value for each station.
#example stats service map, comparing real-time current discharge to history for each site
#reusable for other state(s)
#David Watkins June 2016
library(maps)
library(dplyr)
library(lubridate)
library(dataRetrieval)
#pick state(s) and date
states <- c("WI","MN","ND","SD","IA")
storm.date <- "2016-07-12"
#download each state individually
for(st in states){
stDV <- renameNWISColumns(readNWISdata(service="dv",
parameterCd="00060",
stateCd = st,
startDate = storm.date,
endDate = storm.date))
if(st != states[1]){
storm.data <- full_join(storm.data,stDV)
sites <- full_join(sites, attr(stDV, "siteInfo"))
} else {
storm.data <- stDV
sites <- attr(stDV, "siteInfo")
}
}
#retrieve stats data, dealing with 10 site limit to stat service requests
reqBks <- seq(1,nrow(sites),by=10)
statData <- data.frame()
for(i in reqBks) {
getSites <- sites$site_no[i:(i+9)]
currentSites <- readNWISstat(siteNumbers = getSites,
parameterCd = "00060",
statReportType="daily",
statType=c("p10","p25","p50","p75","p90","mean"))
statData <- rbind(statData,currentSites)
}
statData.storm <- statData[statData$month_nu == month(storm.date) &
statData$day_nu == day(storm.date),]
finalJoin <- left_join(storm.data,statData.storm)
finalJoin <- left_join(finalJoin,sites)
finalJoin[,grep("_va",names(finalJoin))] <- sapply(finalJoin[,grep("_va",names(finalJoin))], function(x) as.numeric(x))
#remove sites without current data
finalJoin <- finalJoin[!is.na(finalJoin$Flow),]
#classify current discharge values
finalJoin$class <- NA
finalJoin$class[finalJoin$Flow > finalJoin$p75_va] <- "navy"
finalJoin$class[finalJoin$Flow < finalJoin$p25_va] <- "red"
finalJoin$class[finalJoin$Flow > finalJoin$p25_va &
finalJoin$Flow <= finalJoin$p50_va] <- "green"
finalJoin$class[finalJoin$Flow > finalJoin$p50_va &
finalJoin$Flow <= finalJoin$p75_va] <- "blue"
finalJoin$class[is.na(finalJoin$class) &
finalJoin$Flow > finalJoin$p50_va] <- "cyan"
finalJoin$class[is.na(finalJoin$class) &
finalJoin$Flow < finalJoin$p50_va] <- "yellow"
#take a look at the columns that we will plot later:
head(finalJoin[,c("dec_lon_va","dec_lat_va","class")])
## dec_lon_va dec_lat_va class
## 1 -92.09389 46.63333 navy
## 2 -91.59528 46.53778 navy
## 3 -90.96324 46.59439 navy
## 4 -90.59000 46.39472 navy
## 5 -90.69630 46.48661 navy
## 6 -90.90417 46.49722 navy
Make the static plot
The base map consists of two plots. The first makes the county lines with a gray background, and the second overlays the heavier state lines. After that we add the points for each stream gage, colored by the column we added to finalJoin. In the finishing details, grconvertXY is a handy function that converts your inputs from a normalized (0-1) coordinate system to the actual map coordinates, which allows the legend and scale to stay in the same relative location on different maps.
#convert states from postal codes to full names
states <- stateCdLookup(states, outputType = "fullName")
par(pty="s")
map('county',regions=states,fill=TRUE, col="gray87", lwd=0.5)
map('state',regions=states,fill=FALSE, lwd=2, add=TRUE)
points(finalJoin$dec_lon_va,
finalJoin$dec_lat_va,
col=finalJoin$class, pch=19)
title(paste("Daily discharge value percentile rank\n",storm.date),line=1)
par(mar=c(5.1, 4.1, 4.1, 6), xpd=TRUE)
legend.colors <- c("cyan","yellow",
"red",
"green","blue",
"navy")
legend.names <- c("Q > P50*","Q < P50*",
"Q < P25",
"P25 < Q < P50","P50 < Q < P75",
"Q > P75")
legend("bottomleft",inset=c(0.01,.01),
legend=legend.names,
pch=19,cex = 0.75,pt.cex = 1.2,
col = legend.colors,
ncol = 2)
map.scale(ratio=FALSE,cex = 0.75,
grconvertX(.07,"npc"),
grconvertY(.2, "npc"))
text("*Other percentiles not available for these sites", cex=0.75,
x=grconvertX(0.2,"npc"),
y=grconvertY(-0.08, "npc"))
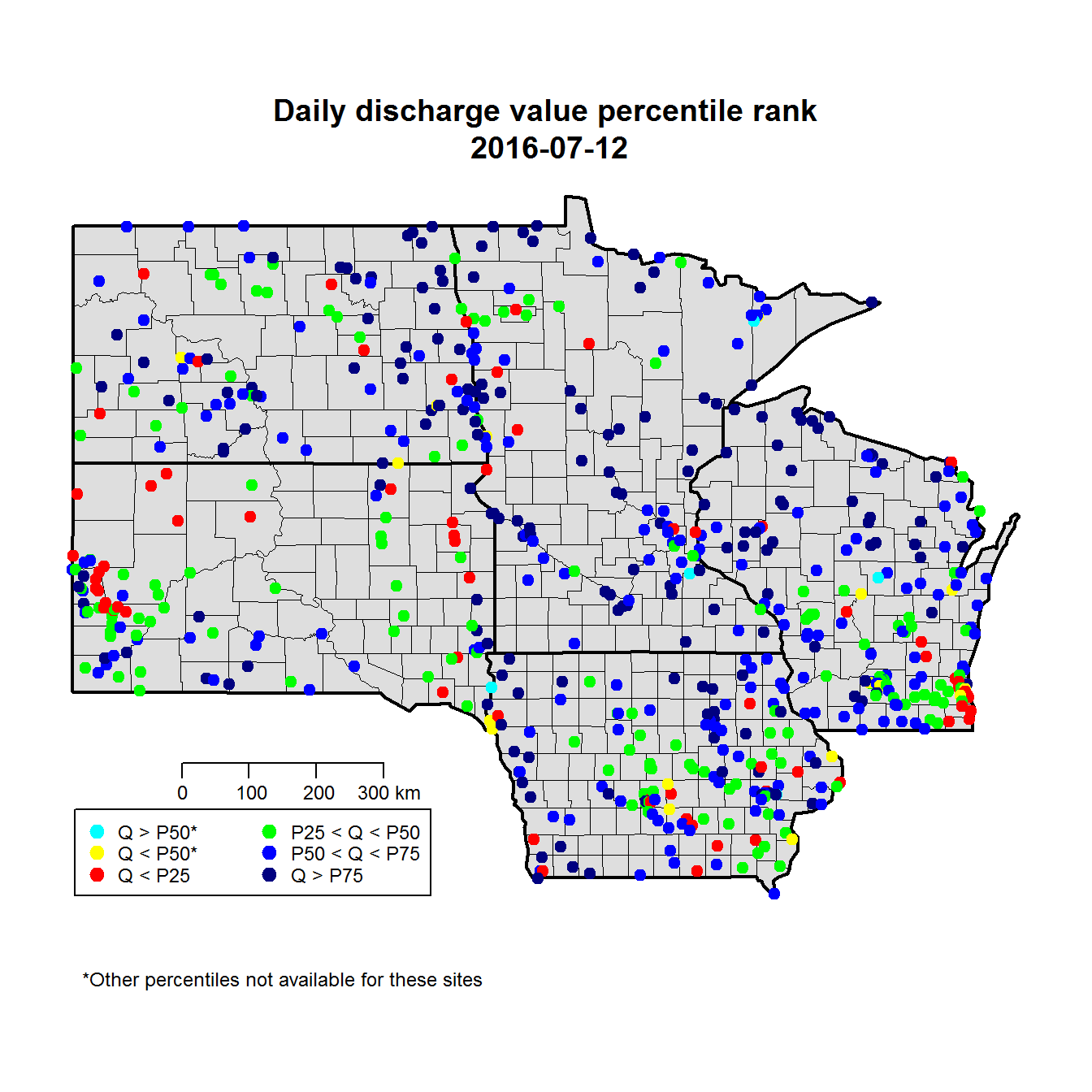
Map discharge percentiles
Make an interactive plot
Static maps are great for papers and presentations. When possible, interactive maps allow the reader more flexibility to examine the data. The R leaflet package makes it easy to create useful interactive maps:
library(leaflet)
finalJoin$popup <- with(finalJoin, paste("<b>",station_nm,"</b></br>",
"Measured Flow:",Flow,"ft3/s</br>",
"25% historical:",p25_va,"ft3/s</br>",
"50% historical:",p50_va,"ft3/s</br>",
"75% historical:",p75_va,"ft3/s"))
leafMapStat <- leaflet(data=finalJoin) %>%
addProviderTiles("CartoDB.Positron") %>%
addCircleMarkers(~dec_lon_va,~dec_lat_va,
color = ~class, radius=3, stroke=FALSE,
fillOpacity = 0.8, opacity = 0.8,
popup=~popup)
leafMapStat <- addLegend(leafMapStat,
position = 'bottomleft',
colors= legend.colors,
labels= legend.names,
opacity = 0.8)
Questions
Please direct any questions or comments on dataRetrieval to: https://github.com/DOI-USGS/dataRetrieval/issues
Categories:
Keywords:
Related Posts
Easy hydrology mapping with nhdplusTools, geoconnex, and ggplot2
November 28, 2025

Go from hard-to-read default visuals to easy-to-read river maps in a few easy steps!
Tutorial of dataRetrieval's newest features in R
November 26, 2025
This article will describe the R-package
dataRetrieval, which simplifies the process of finding and retrieving water from the U.S. Geological Survey (USGS) and other agencies. We have recently released a new version ofdataRetrievalto work with the modernized Water Data APIs . The new version ofdataRetrievalhas several benefits for new and existing users:Formatting guidance for USGS Samples Data Tables
May 6, 2025
Recently, changes were made to the delivery format of USGS samples data (learn more here ). In this blog, we describe the impact to users and show an example of how to use R to convert WQX-formatted water quality results data to a tabular, or “wide” view.
Reproducible Data Science in R: Iterate, don't duplicate
July 18, 2024
Overview
This blog post is part of a series that works up from functional programming foundations through the use of the targets R package to create efficient, reproducible data workflows.
Reproducible Data Science in R: Writing better functions
June 17, 2024
Overview
This blog post is part of a series that works up from functional programming foundations through the use of the targets R package to create efficient, reproducible data workflows.

